EecSeq Bioinformatics
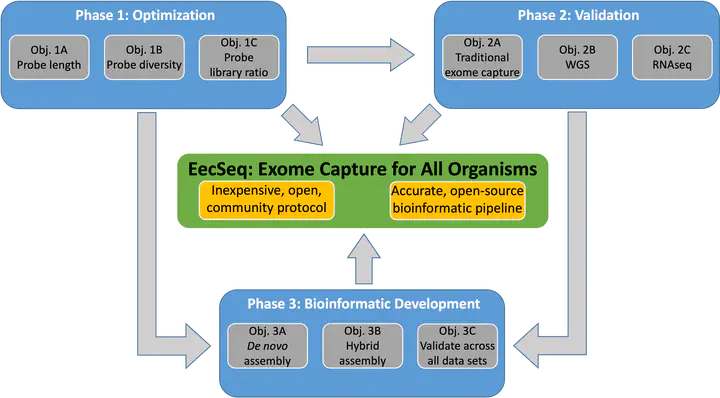
To develop EecSeq into an exome capture method for any organism, the accompanying bioinformatics pipeline needs to be capable of de novo assembly of exon loci directly from captured genomic reads. Assembling exon loci is a new bioinformatic challenge as traditional exome capture relies on designed probes from genomic and/or transcriptomic data. Accurate de novo assembly is critical to population level inference derived from reduced representation data sets, and errors, artifacts, and biases in assembly are still problematic in both RADseq and RNAseq. Leveraging the chromosome-level assembly of the eastern oyster genome, we are developing two complementing de novo assembly methods for EecSeq: one utilizing only captured genomic reads and a second hybrid method that will utilize sequences from the cDNA probes (when sequenced) and captured genomic reads. The final output will be an open source bioinformatics pipeline for EecSeq.
